Announcing the Targeted Loci Database for 16S and other amplicon sequencing
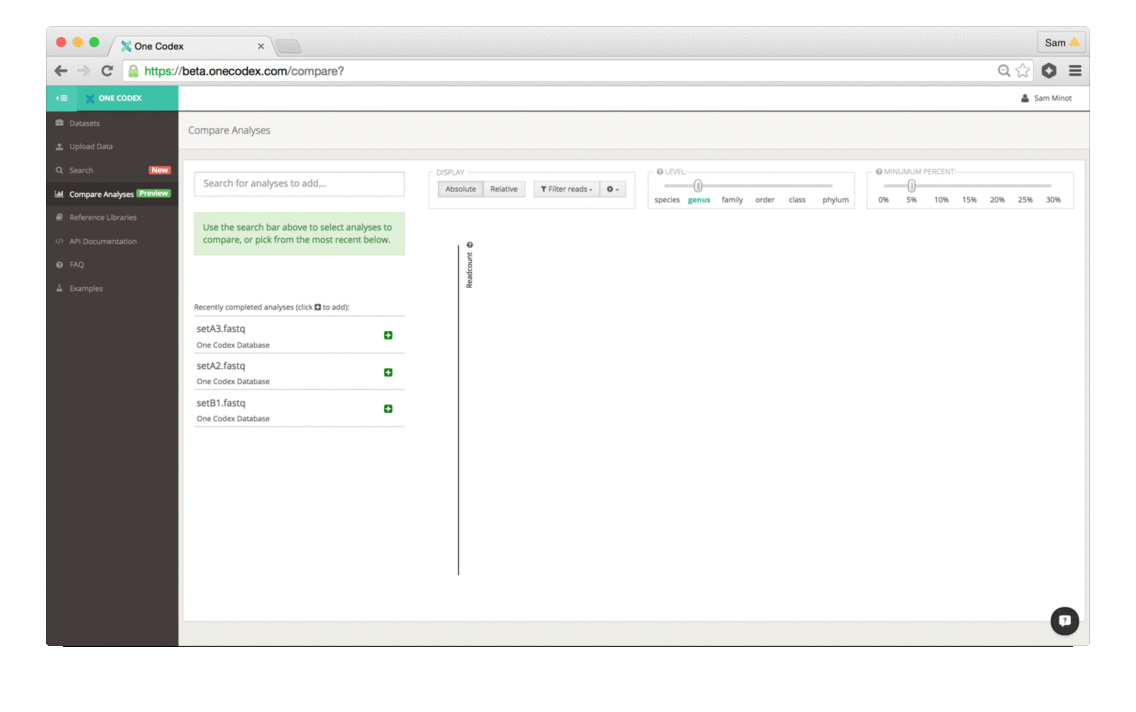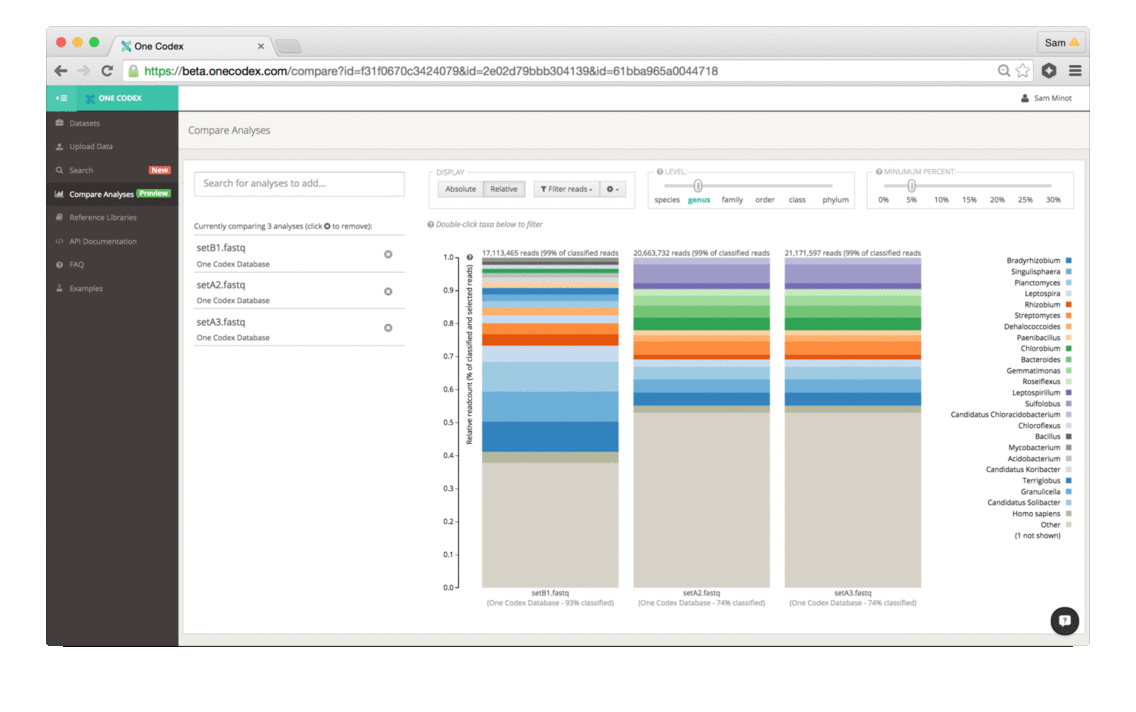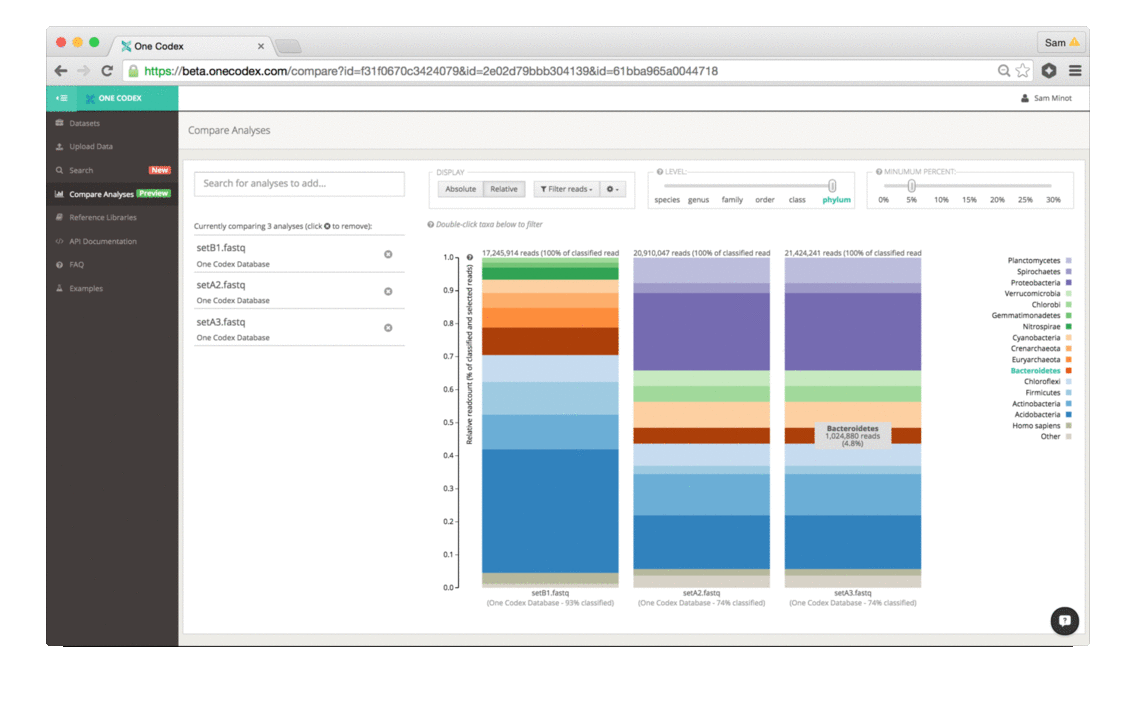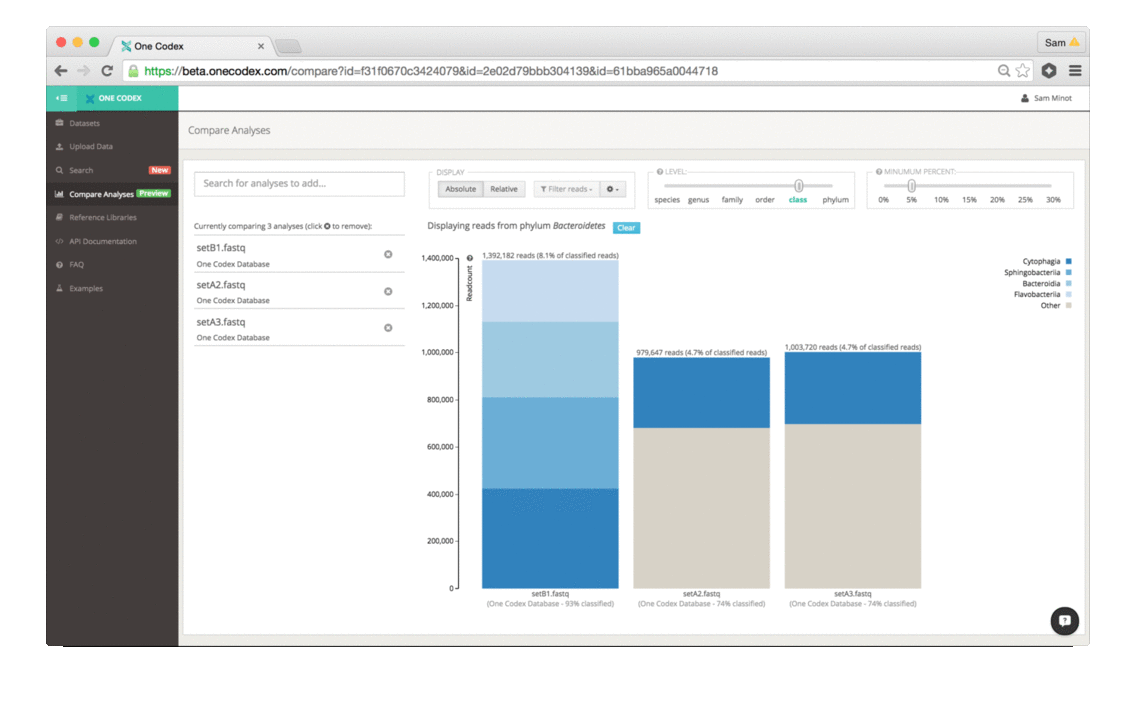
Scientists that study the microbiome generally use two different methods to analyze samples – sequencing all of the DNA in a sample (whole genome sequencing) or targeting a specific marker gene (e.g., 16S, 18S, ITS). While whole genome sequencing (WGS) enables high-resolution taxonomic and functional characterization of microbiome samples, 16S sequencing is a cost effective technique for broad community surveys across large numbers of samples. Today, we’re excited to announce that One Codex is launching a powerful new tool for 16S and other amplicon sequencing – making high-quality, reference-based analysis of marker gene studies easier, faster, and more accessible.