Notebooks (and more!)
Today, we’re extremely excited to announce several new features we’ve been working on for the past few months. Collectively, these should make it both easier and faster to perform custom, large-scale analyses, explore your data, and build applications atop the One Codex platform:
- A new easier-to-use, more powerful API (read the docs)
- A new version of our command-line interface and an accompanying Python client library for quickly getting started with the new API (take a peek on Github)
- And last but not least – interactive notebooks built directly into the platform!
The new Notebooks feature allows you to launch secure Jupyter (née IPython) notebooks automatically configured for access to your samples and analyses on One Codex. Internally, we’ve already found these incredibly useful for both quick, flexible explorations of metagenomic data as well as more sophisticated, formal analyses.
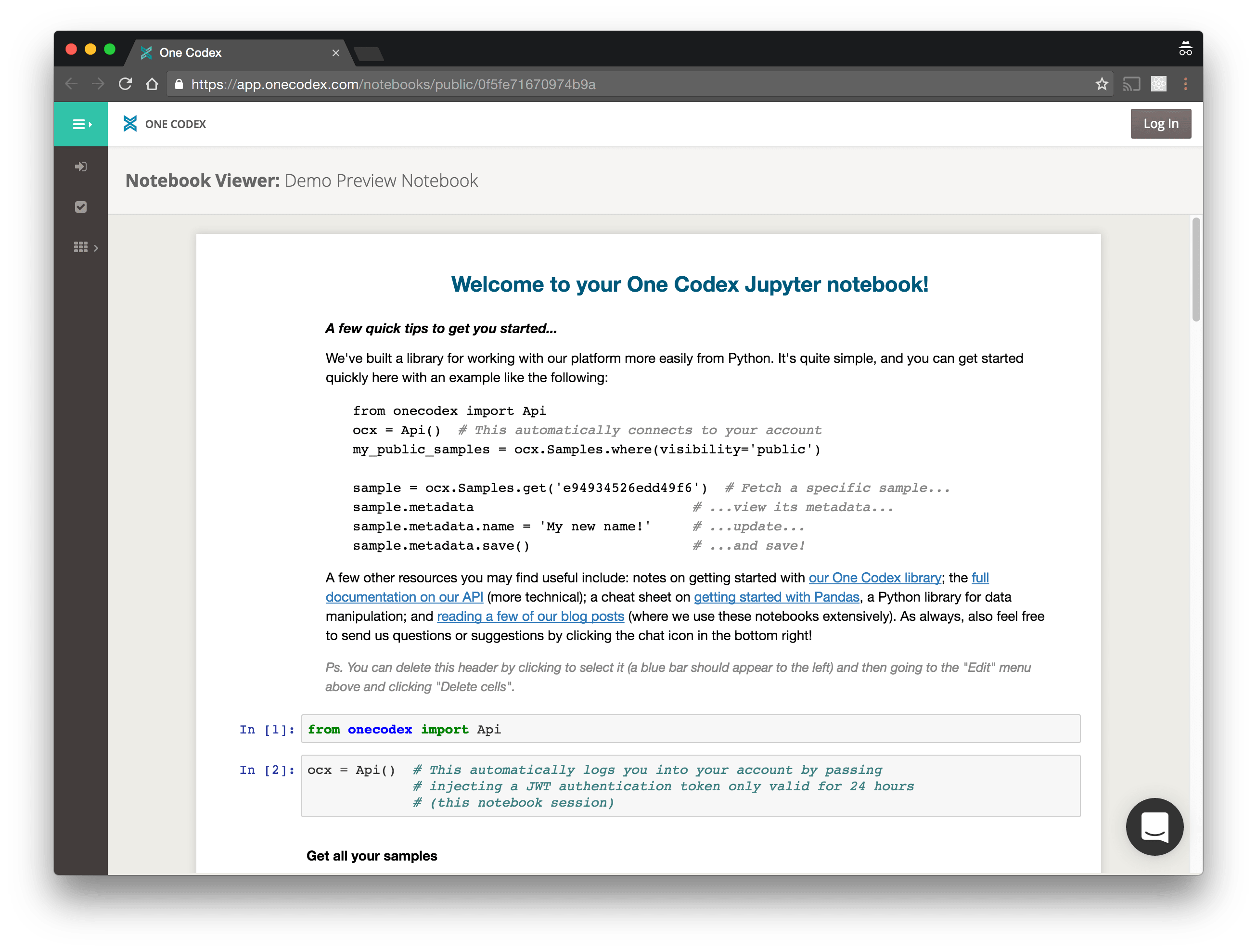
Notebook details
Availability: Notebooks are in beta and available for all users. We would love to hear your feedback as you start using notebooks in your analyses.
Technical specifications: Our default notebooks provide 1 CPU and 4GB of RAM. Notebooks run in a secure, multi-tenant environment and are protected by your One Codex login credentials. There are currently no maximum runtime limits imposed on notebooks, but they will be saved and shut down if they are idle and have not been accessed in the last 4 hours.
Pricing: Basic notebook access will be free. If you need notebooks with additional resources or would like to run your notebooks on dedicated infrastructure, please send us a note.
Questions & feedback
We’re really excited to see what folks build using the new notebooks and, as always, are happy to answer any questions and grateful for any feedback you have as you play around with them.