Cautious Metagenomics: A Story of Anthrax in the NYC Subway
Ok, anthrax might sound a bit scary, but this is a story about something that should make you feel good.
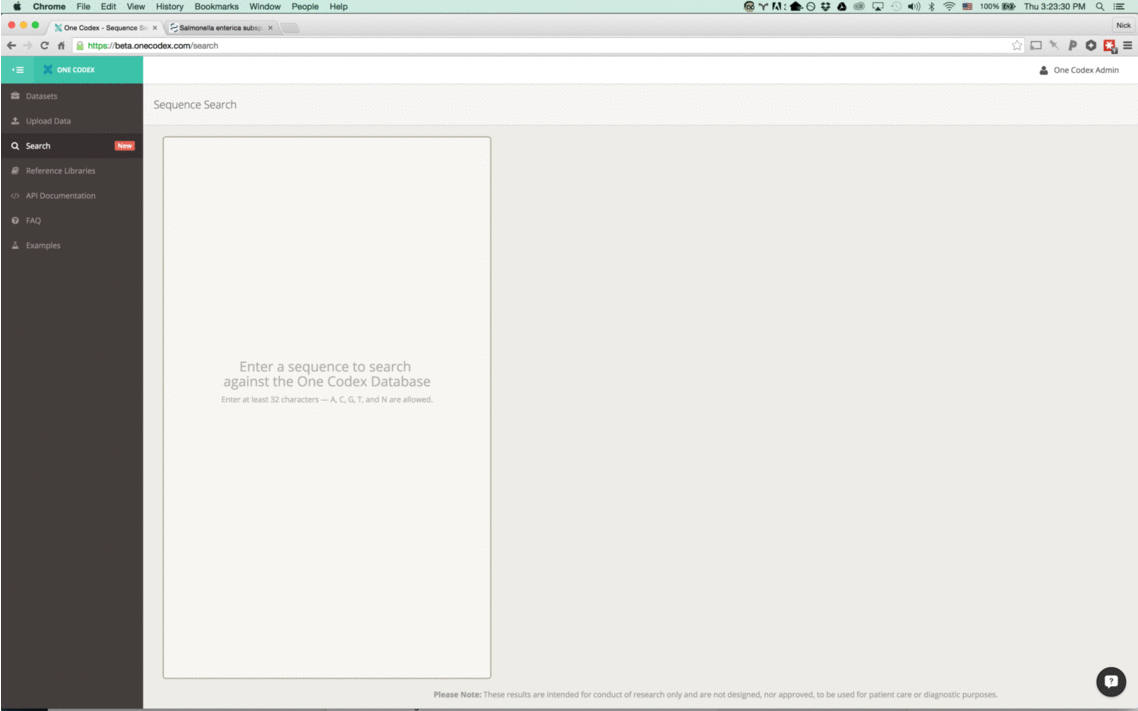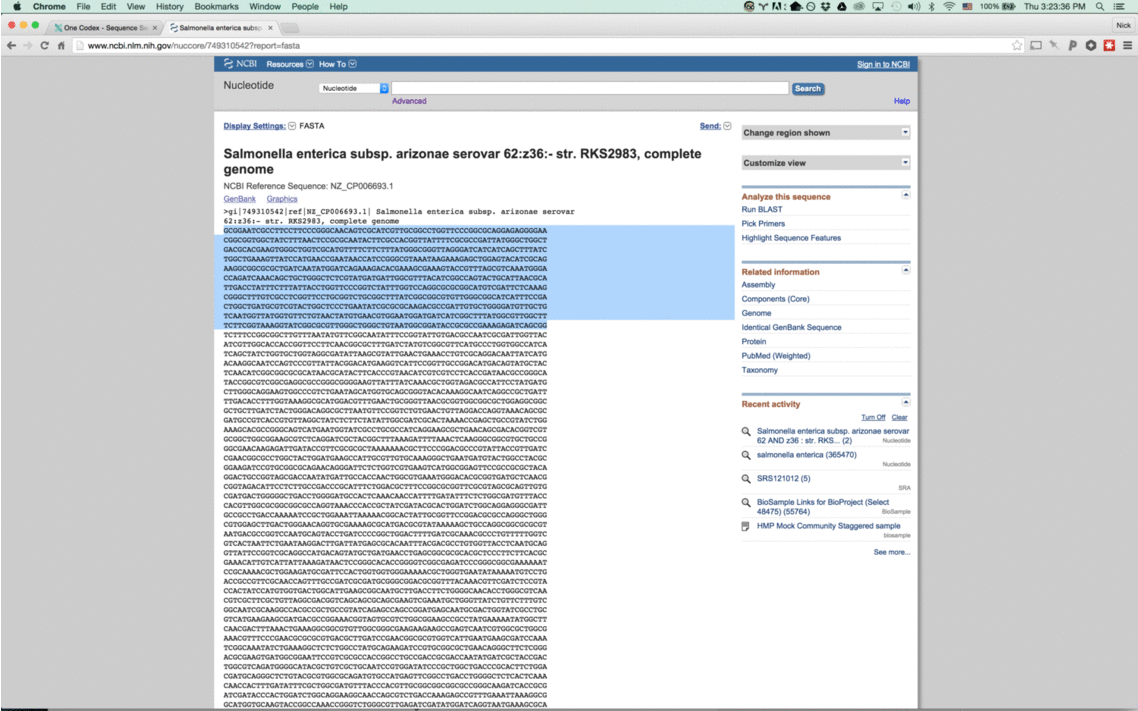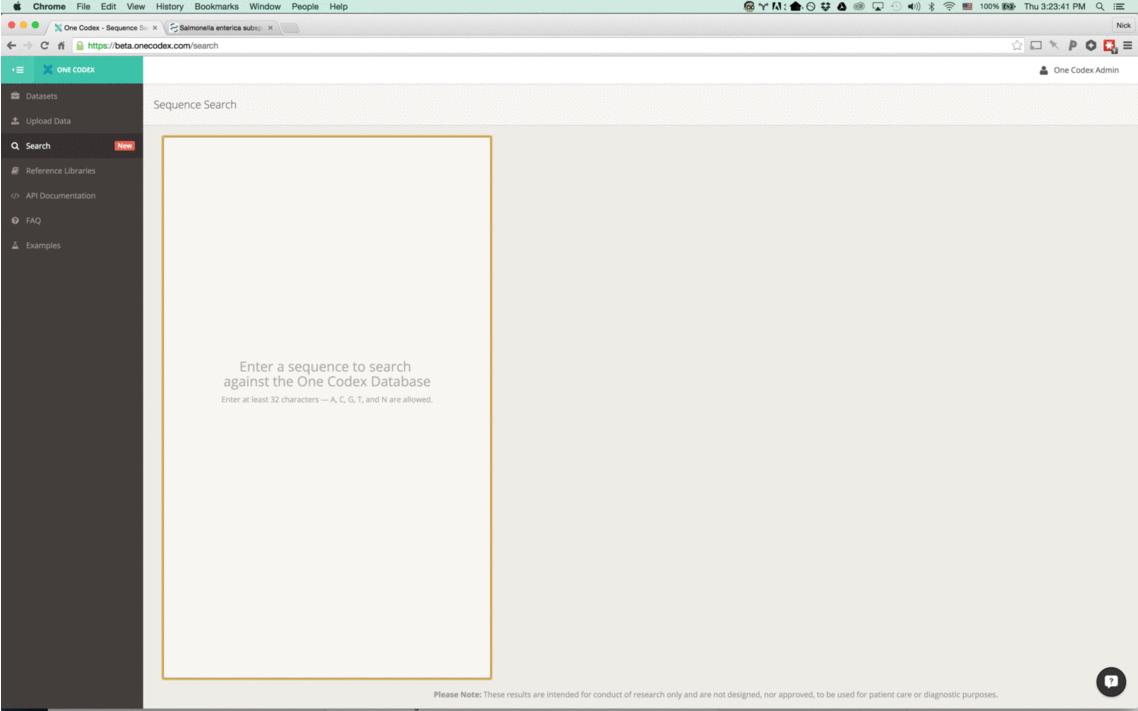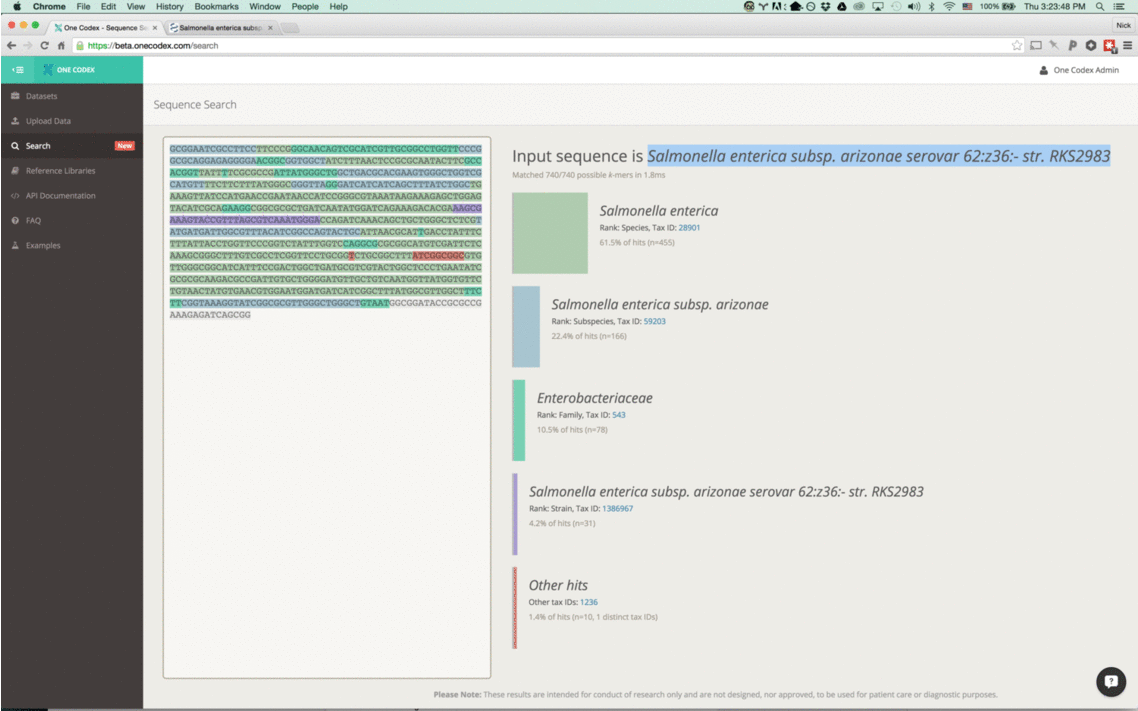
Wait, is that really Anthrax?
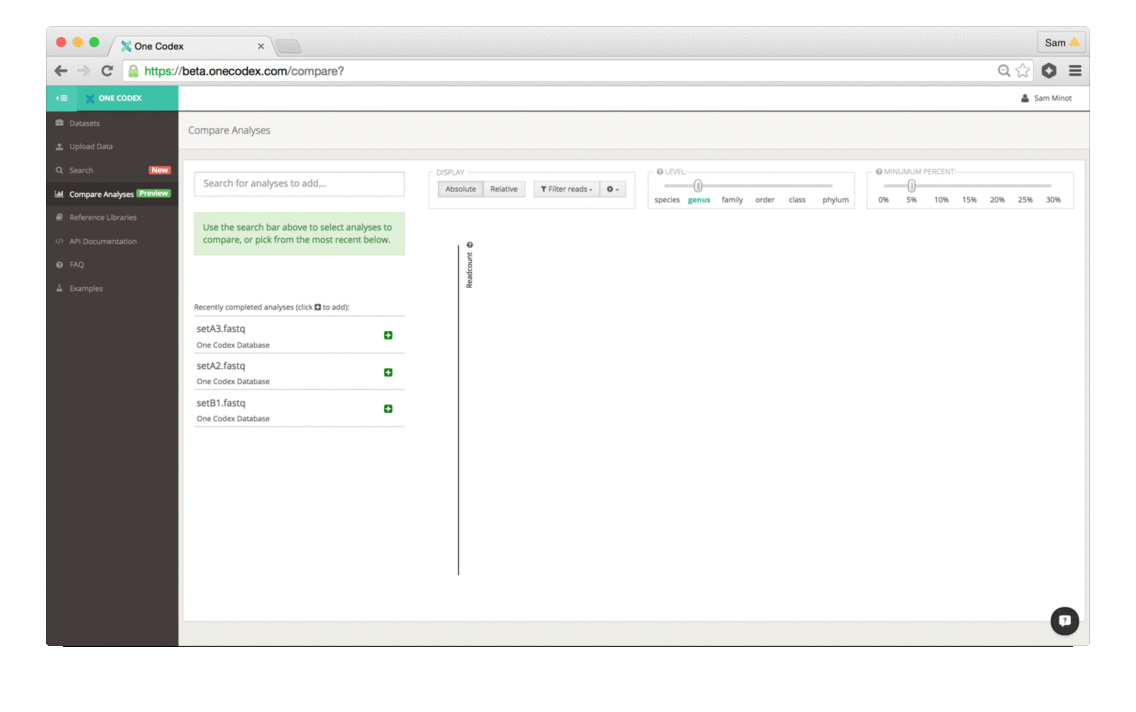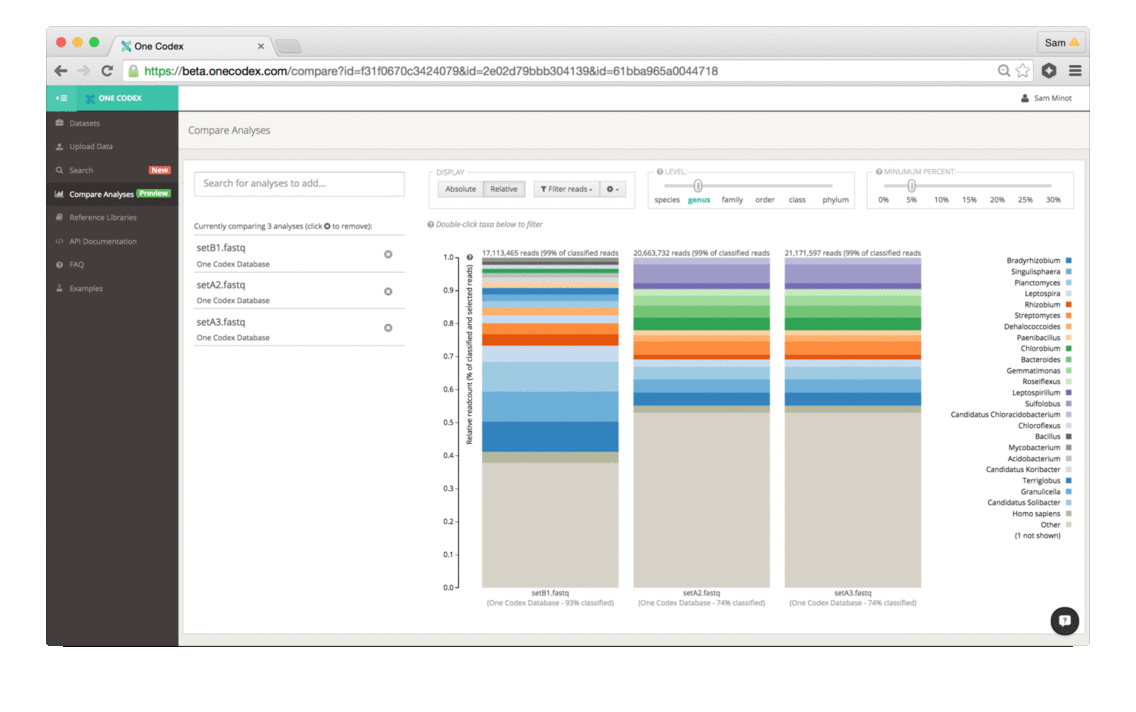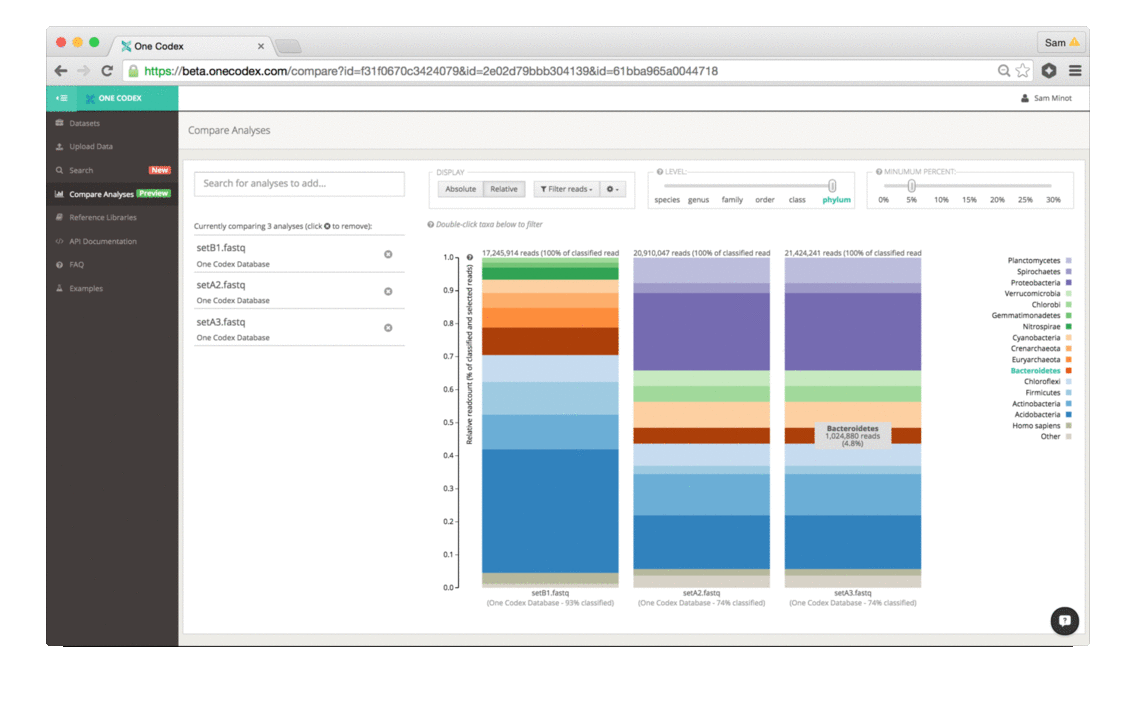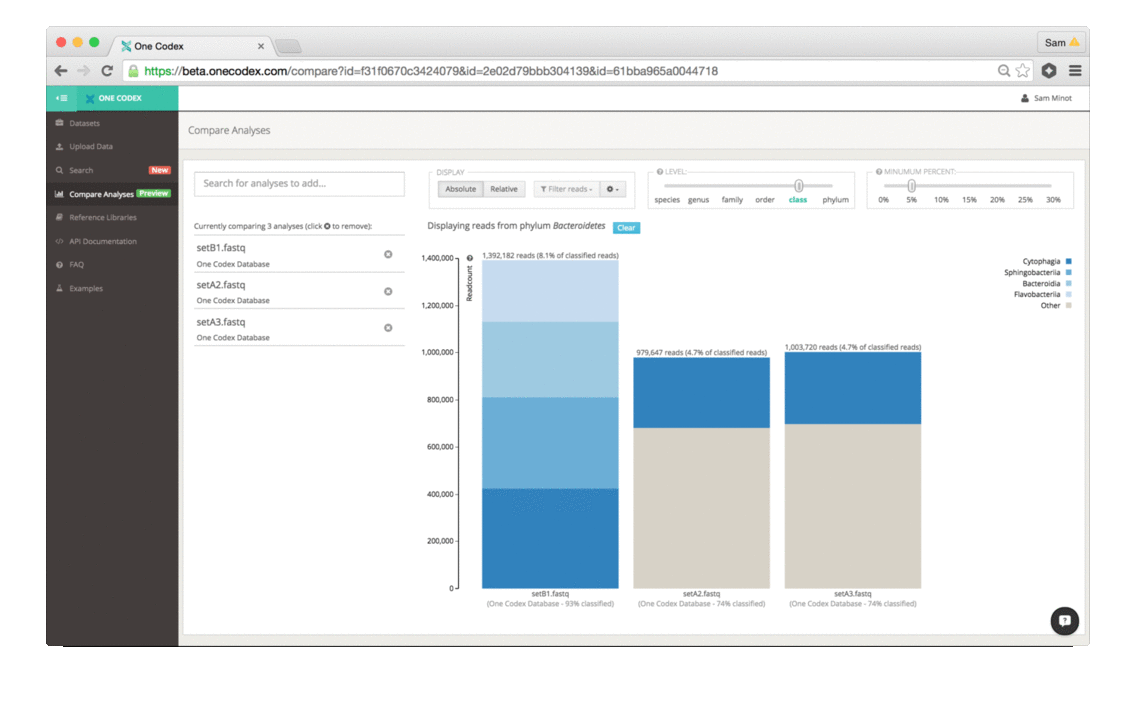
Back in the Spring of 2015, researchers studying the microbial ecology of the built environment generated a very large amount of genomic data from microbes found in the New York City subway system. To give you some idea of the magnitude, they generated 10.4 billion sequence reads across 1,457 samples. That’s a lot of microbiome data.