A new and improved pipeline for detecting antimicrobial resistance markers in metagenomic data
In clinical settings, detecting antimicrobial resistance (AMR) markers directly from metagenomic data is not only becoming increasingly commonplace but necessary for human health and safety. This shift is largely driven by a “looming crisis” in which the emergence and spread of resistant pathogens (often a result from antibiotic overuse) are outpacing the development of new antibiotics [1] [2]. In response to this urgent challenge, there has been a substantial proliferation of useful tools designed to rapidly and accurately detect AMR markers [3]. These tools have the potential to provide actionable insights for clinicians within hours, rather than days; coupled with current diagnostic methods, they can help inform treatment decisions and prevent the misuse of antibiotics. Here at One Codex, we are committed to staying at the forefront of these advancements and providing our users with the appropriate metagenomic tools for use in research and clinical settings. In the past, we have provided our users with the tools to identify resistance in foodborne pathogens, we have released free-for-use pipelines for SARS-CoV-2 variant detection and processing, and today we are excited to introduce our new and improved AMR detection pipeline.
Figure 1. Treemap depicting the distribution of AMR sequences across antimicrobial classes and resistance mechanisms. AMR sequences are broadly colored according to their antimicrobial type (e.g., drugs, biocides, etc.) and grouped according to their chemical class (e.g., betalactams, glycopeptides, etc.). Numbers indicate the amount of AMR sequences belonging to a given chemical class.
Our new AMR detection pipeline and database features over 8,500 distinct AMR sequences, an order of magnitude more than our previous database! [Fig. 1] Based on the MEGARes resource and ontology, our new AMR pipeline covers a wide variety of antimicrobial classes and resistance mechanisms [4]. However, in many cases simply detecting the presence of an AMR gene is not enough. Often, resistance is conferred by single nucleotide polymorphisms (SNPs) or other mutations in the genome [5]. For the relevant markers, we also detect the presence of these SNPs and provide additional information such as alignment statistics.
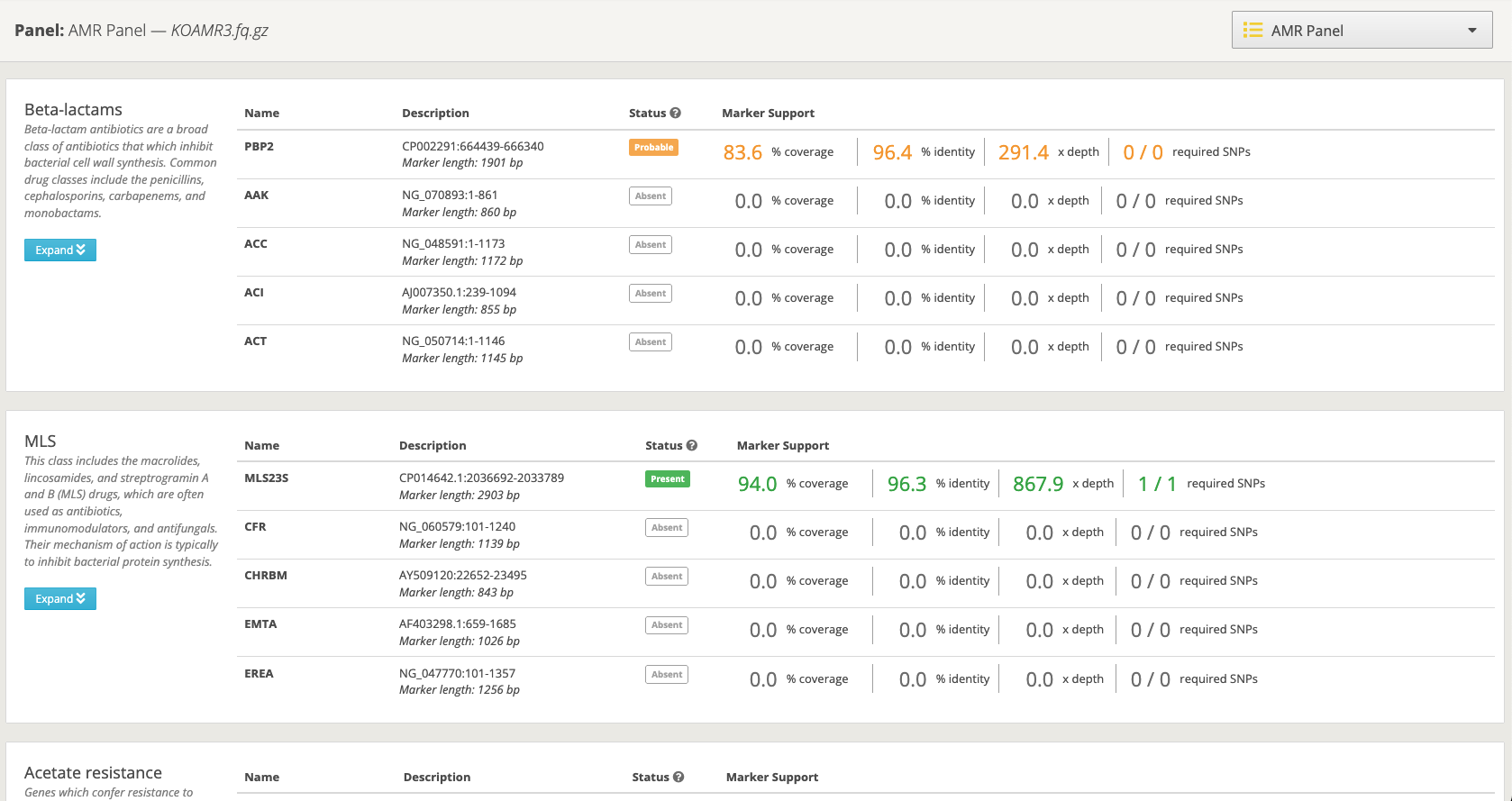
Figure 2. Screenshot of the new, updated AMR panel.
This updated AMR detection pipeline is now available to all users and ready to run on all samples. Like our other workflows and analyses, you can find it under the “Run Workflows” page. In addition to the classic results page we also provide a downloadable PDF report and offer raw pipeline outputs for any microbiologist or bioinformatician who wants to dive deeper into their data.
References
- Aslam, B. et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist 11, 1645–1658 (2018).
- Brown, E. D. & Wright, G. D. Antibacterial drug discovery in the resistance era. Nature 529, 336–343 (2016).
- Mendes, I. et al. hAMRonization: Enhancing antimicrobial resistance prediction using the PHA4GE AMR detection specification and tooling. 2024.03.07.583950 Preprint at https://doi.org/10.1101/2024.03.07.583950 (2024).
- Bonin, N. et al. MEGARes and AMR++, v3.0: an updated comprehensive database of antimicrobial resistance determinants and an improved software pipeline for classification using high-throughput sequencing. Nucleic Acids Res 51, D744–D752 (2022).
- Leanse, L. G., Marasini, S., dos Anjos, C. & Dai, T. Antimicrobial Resistance: Is There a ‘Light’ at the End of the Tunnel? Antibiotics (Basel) 12, 1437 (2023).