Improved SARS-CoV-2 Variant Detection with One Codex
In March of 2020, a few weeks after the WHO declared a global COVID-19 pandemic, One Codex released a free, publicly available pipeline for processing different types of SARS-CoV-2 sequencing data (WGS, amplicon, hybrid capture). This pipeline is automatically run on any One Codex sample in which SARS-CoV-2 is detected, and is part of our effort to contribute what we can towards the public health response.
We’re happy to announce that One Codex has now updated our SARS-CoV-2 pipeline with additional and improved features that reflect the changing nature of the pandemic. The pipeline now deals exclusively with tiling amplicon data generated by ARTIC V3 protocols, which have been widely adopted for detecting SARS-CoV-2 variants. The pipeline takes in either Oxford Nanopore or Illumina FASTQ data and processes them as follows:
- Map reads to the Wuhan-Hu-1 reference and trim ARTIC primer sequences
- Generate a consensus sequence (bcftools for Illumina; medaka for Oxford Nanopore)
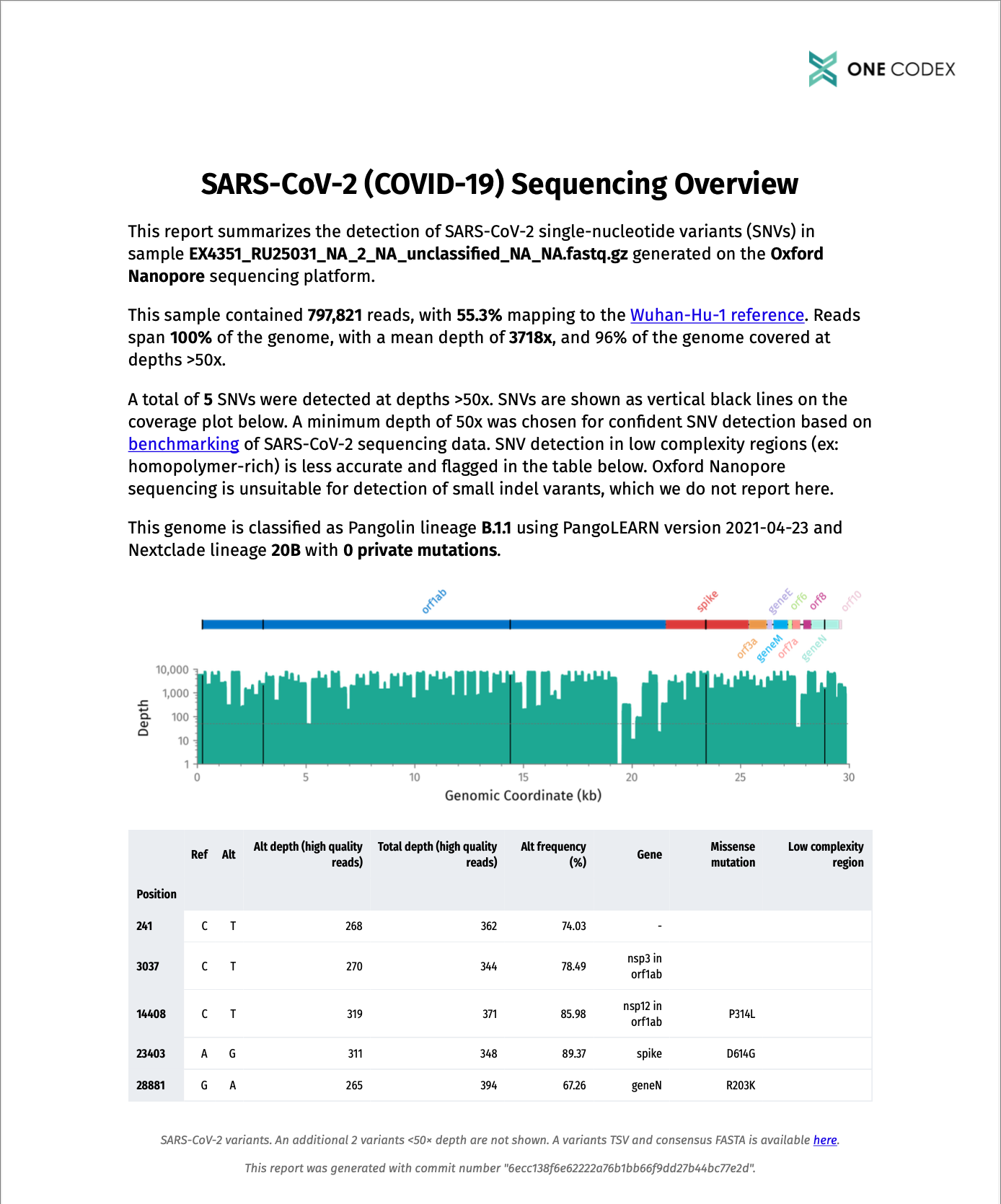
- Call variants with a limit of 200x coverage, as recommended by the ARTIC network. While indels and SNVs are reported for Illumina data, only SNVs are reported for Oxford Nanopore based on benchmarking studies that indicate small indel detection is unreliable
- Assign Pangolin and Nextclade lineages
- Predict amino acid mutations
- Predict consequences of compound variants (ex: adjacent SNVs on the same codon; frame-shifting indels followed by frame-restoring indels)
Rigorous quality checks are implemented throughout the pipeline, including flagging of variants in low complexity regions for error-prone Oxford Nanopore data and conservative lineage calls (no lineage assignments will be reported if the consensus sequence has too many N’s or is too fragmented).
A PDF report is generated for each sample that tells you at a glance whether primer dropout has occurred, which amino acid mutations are present, and whether the sample contains a variant of concern.
Examples in the wild:
Multiple spillovers and onward transmission of SARS-CoV-2 in free-living and captive white-tailed deer https://doi.org/10.1101/2021.10.31.466677